Deathbase is a database of proteins involved in different Cell Death processes. It is aimed to compile relevant data on the function, structure and evolution of this important cellular proccess in several organisms. Information contained in the database is subject to manual curation. This page provides information on how the database is developed and some basic help on how to access and query the database. If you need additional help please contact us.
- Accessing the database
- Proteins curated by DeathBase
- Curation of the database and wiki-based annotation system.
- Homologs to curated proteins.
- Cell death Processes and Pathways.
- Functional information.
- Structural information.
- Evolutionary information.
- Information on gene regulation.
- Methods used in the database.
Accessing the database:

Through the top bar the home page provides direct access to the different sections of the database:
- Proteins: allows browsing the lists of proteins in different species;
- Pathways: allows searching for proteins involved in specific cell death pathways;
- Families: allows complex searches for proteins belonging to specific structural families or having a particular combinations of domains;
- Downloads: allows downloading database information in a tabular format
- Links: provides links to useful resources;
- About: information about database curators and developers;
- WIKI: provides access to community-based annotations; 8) Help: provides help on the use of the database.
Quick search for a protein entry:
The quick database access box provides an easy way for querying the database. Just enter the protein name or identifier (ENSEMBL, Swissprot). You can restrict the search to a specific species by selecting the relevant species.
Proteins curated by DeathBase
One of the aims of this database is to eliminate “noise” in the field by focusing only on the proteins which play direct roles in cell death, rather than including every protein whose overexpression induces apoptosis. We included in the DeathBase proteins that fit in one or more of the following categories: i) which are part of the core evolutionarily conserved machinery of apoptosis (essentially, caspases, Bcl-2 family members, Apaf-1/FADD-like adapter proteins, IAPs, death receptors and ligands, and proteins involved in phagocytosis of apoptotic corpses); ii) proteins that directly regulate other proteins of the core machinery of apoptosis (for example, Granzyme B or IAP antagonists); iii) proteins that contain domains involved in apoptotic interactions such as CARDS or Death Domains; iv) proteins that participate in non-apoptotic forms of cell death (for instance, RIP or Drosophila autophagic proteins); v) that are homologous to central mediators of apoptosis, even if their role in apoptosis has been discounted or is unproven (for instance, non-apoptotic caspases). In these latter cases, we aim to provide comments and references about their physiological roles. Conversely, for the sake of clarity, DeathBase does not include proteins that are not central to the apoptotic process, even if they may play a role on initiation of a cell death pathway. Among these, we did not include: i) receptors for survival factors, such as IL-3 or NGF receptors; ii) proteins that participate in a specific signaling pathway that eventually may end up in induction or activation of apoptotic proteins, such as transcription factors or stress-signaling proteins; iii) most caspase substrates (for a database on caspase substrates please check CASBAH database).
DeathBase includes proteins involved in cell death in five model species: Human, mouse, zebrafish, fly and worm. These proteins are labeled as “manually curated”. The choice of these five species is based on their extensive use as model organisms for the study of apoptosis and the fact that most studies describing the experimental characterization of key apoptotic players are circumscribed to these organisms.
Curation of the database and wiki-based annotation system
A team of database curators has been set up with the participation of experts in cell death in a specific model organism or protein family. A password protected entry web-editor has been implemented that allows authorized curators to directly add new entries or add and modify relevant information in the database entries. Additionally, a wiki-based community annotation page is provided for each entry in which general users can add comments. Similar to the wikipedia, this resource will allow continuous expansion of the information contained in the database. We encourage users of the DeathBase worlwide to use this resource and contribute to our understanding of the fascinating process of cell death.
Homologs of curated proteins
We searched for homologs of curated proteins in the other model species and a total of 23 non-reference species. These proteins are included in the database by virtue of their homology relationships and are therefore labeled as “homology based”, in opposition to “manually curated” proteins. The reason for including these homology-based proteins in other species is manifold. First, there is a growing interest in studying apoptosis in alternative model organisms, since each species might be particularly suited to a specific type of experiments. Second, comparative analyses across different eukaryotic phyla serve to gain insight on the evolution of this biological process. Finally, comparative genomics might be used to predict the function of partially characterized apoptotic proteins or to find novel components of the pathway18. Both manually curated and homology-based entries are populated with extensive information on their structure, function and evolution, based on data extracted from external repositories or our own analyses. Below we will describe the main features.
Cell Death Processes and Pathways
Apoptotic proteins are classified according to the pathway in which they participate: extrinsic or intrinsic; pre-mitochondrial signaling events or post-mitochondrial caspase activation, and apoptotic cell clearance. Proteins that participate in other, non-apoptotic forms of cell death have been assigned the descriptor “cell death (other)”. A few examples of these non-apoptotic cell death processes are: autophagic cell death during Drosophila development, pyroptosis and cornification. Death-receptor induced necrosis --also called necroptosis--, has been included as a separate cell death process descriptor, because at this point there is a large number of proteins that have been shown to participate in this process. Additionally, we have created a fourth label that applies to proteins with immune function. The reason is that many caspases, caspase adapter proteins and IAPs are not involved in apoptosis, but they participate in immunity, both in mammals and insects.
Functional information
Known functions of the proteins are indicated in the curator comments section of each entry. A special emphasis is devoted to the function of that protein within programmed cell death pathways but we also refer to alternative roles in other pathways. Additionally, all Gene Ontology terms associated to that protein are listed. These terms are structured descriptions of the molecular function of the protein, its cellular localization and the biological processes in which it is involved. Links to the Gene Ontology database allows users to get additional information on these descriptors and to find other proteins with similar annotations. The pathways in which a given protein is playing a role can be also explored by linking from DeathBase to the pathways included in the KEGG20 database. Additional links are provided to the respective model organism databases for proteins encoded in the reference genomes: Mouse Genome Information21 for Mouse, ZFIN for zebra fish, FlyBase for Drosophila melanogaster and wormbase for Caenorhabditis elegans. Additionally, whenever possible, entries are linked to other general databases such as Uniprot, Ensembl, Wikigenes (www.wikigenes.org), Genecards, and PIR.
Structural information.
DeathBase provides structural information for each protein in various ways. First, the apoptosis-related proteins in the database are classified into the structural families defined by Reed et al., attending to the presence of apoptotic domains: Bcl-2 (which includes BH3-containing proteins), Caspase, Death Domain, Death Effector Domain, CARD (Caspase Recruitment Domain) and BIR (Baculovirus IAP repeat). The domain composition of a protein is described by mapping its sequence to domains and motifs in PFAM, Interpro Prosite and SMART databases. When available, links to the protein 3D structures in PDB are provided. Possible physical interactions with other proteins can be obtained through IntAct, a database providing interacting protein pairs from small and large-scale experiments.
Evolutionary information
Setting all apoptotic proteins within an evolutionary framework is a central goal of DeathBase. Therefore we performed extensive homology searches in eukaryotic organisms for all curated proteins (see Material and Methods). Additional homologs in other species can be found by the user by perfoming an easy, one-click Blast search on NCBI or EBI databases through the link provided next to the sequence of the protein. Multiple sequence alignments and Maximum Likelihood phylogenetic trees including homologs from the 28 species surveyed can be easily viewed interactively or downloaded for further processing (see Materials and Methods). Finally, additional phylogenetic information can be retrieved through the links to phylomeDB, a database providing additional phylogenetic trees and alignments as well as phylogeny-based, prediction of duplications and orthology relationships in a broad range of organisms.
Information on regulation
For all reference species, links to ArrayExpress with information on the expression profiles of the genes are provided. Moreover, information on whether the expression of a gene encoding an apoptotic protein might be regulated by small RNAs is provided through links to miRBase.
Methods used in the database
Sequence data
Sequences where downloaded from ensemble release 53, which includes data from 28 fully-sequenced genomes, including Aedes aegypti, Anolis carolinensis, Anopheles gambiae, Bos taurus, Caenorhabditis elegans, Canis lupus familiaris, Ciona intestinalis, Danio rerio, Drosophila melanogaster, Equus caballus, Gallus gallus, Gasterosteus aculeatus, Gorilla gorilla, Homo sapiens, Macaca mulatta, Monodelphis domestica, Mus musculus, Ornithorhynchus anatinus, Oryctolagus cuniculus, Oryzias latipes, Pan troglodytes, Pongo pygmaeus, Rattus norvegicus, Saccharomyces cerevisiae, Taeniopygia guttata, Takifugu rubripes, Tetraodon nigroviridis and Xenopus tropicalis.
Homology searches
Proteins with experimental evidence of being involved in apoptotic pathways in the reference species (Human, Mouse, Zebrafish, Drosophila, and Worm) were used as a query in Blast searches against the rest of the genomes (e-value <10-5 and overlap cut-off of 35%, using low complexity filter).
Phylogenetic reconstruction
Sets of homologous proteins were aligned with MAFFT39 v6 using the L-INS-i option that implements an iterative refinement incorporating local pair-wise alignment information. Maximum Likelihood trees were derived using JTT as an evolutionary model as implemented in PhyML aLRT version40. In all cases, a discrete gamma-distribution model with four rate categories plus invariant positions was used, estimating the gamma parameter and the fraction of invariant positions from the data. Support for the different partitions was computed by approximate likelihood ratio test as implemented in PhyML aLRT, using the option “Minimum of SH-like and Chi2-based”.
Entry page:
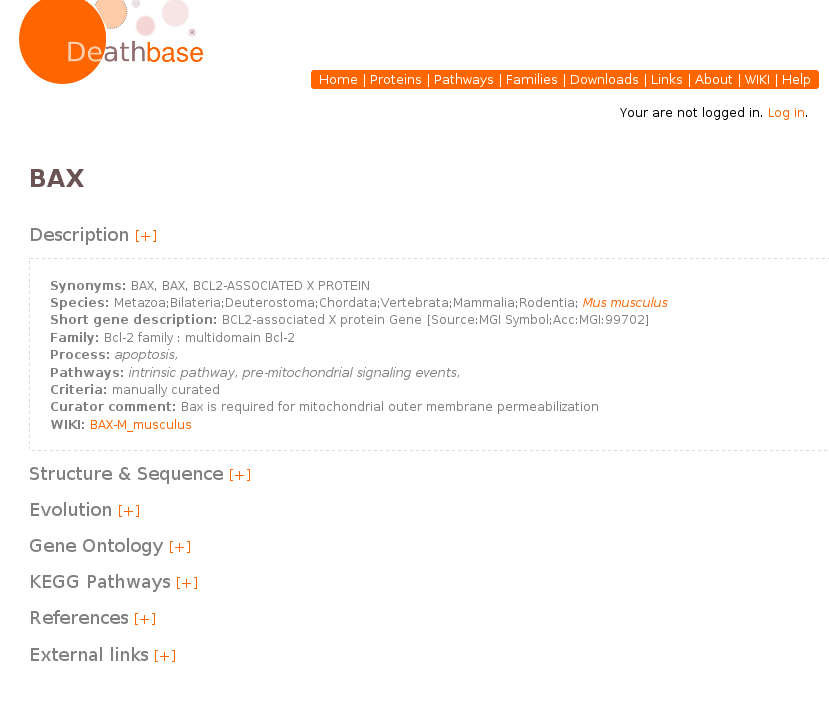
Once you are in the entry page of the desired protein, you will have access to all the relevant information, which is organized into different sections. Click on the “[+]” symbol to expand all the information. Description section (shown in full in this example) provides relevant information on the source of the protein, the curator comments and its assignment to specific a process and pathway. A link to the DeathBase wiki provides access to community based annotations. Additional sections (not displayed in full in this example) provide information on:
- Structure and Sequence of the protein;
- Evolution, including phylogenetic trees and alignments;
- Gene Ontology terms associated to that protein;
- KEGG pathways associated to that entry;
- References
- External links
Families:
- BIR-containing protein.
BIR-containing proteins contain a Baculovirus IAP Repeat. This group includes Inhibitor of Apoptosis Proteins (subfamily IAPs) and proteins with other functions not related to caspase inhibition, for instance, Survivin (subfamily "other"). - IAP. This group includes Inhibitor of Apoptosis Proteins (subfamily IAPs).
- other. Proteins with other functions not related to caspase inhibition, for instance, Survivin (subfamily "other").
- Death receptor.
These proteins are part of the TNF receptor superfamily, and contain a death domain (vertebrates) or transduce death signals (Drosophila Wengen). - Death ligand.
These proteins are part of the TNF family and are ligands for death receptors. - Bcl-2 family.
These proteins contain regions of homology to Bcl-2; either only the BH3 domain (subfamily BH3-only) or other additional domains (subfamily multidomain Bcl-2). - BH3-only. Subfamily BH3-only
- multidomain Bcl-2. Subfamily multidomain Bcl-2
- Caspase.
This family includes proteins of the caspase family and proteins with strong homology such as FLIP. It does not include metacaspases or paracaspases. - IAP antagonist.
IAP antagonists are proteins which have been shown to bind and inhibit IAP function. Examples are Grim and Hid (Drosophila) and Smac/DIABLO (mammals). - Death Domain-containing protein.
Death Domain-containing proteins includes all proteins with this domain, excluding death receptors. - DD adapter protein . The subfamily DD adapter protein comprises proteins such as FADD that contain a Death Domain and function to activate caspases.
- other. The subfamily "other" includes all Death Domain containing proteins with no clear function as caspase activator.
- Death Effector Domain-containing protein.
Death Effector Domain-containing proteins includes all proteins with this domain, excluding caspases. - DED adapter protein. The subfamily DED adapter protein comprises proteins such as FADD that contain a Death Effector Domain and function to activate caspases. The subfamily "other" includes all.
- other. Death Effector Domain containing proteins with no clear function as caspase activator.
- CARD-containing protein.
The family of CARD-containing proteins includes all proteins with this domain except for caspases and IAPs. - CARD adapter protein. The subfamily CARD adapter protein comprises proteins such as Apaf-1 or NALP3 which can activate apoptotic or inflammatory caspases.
- other. The subamily "other" includes all CARD-containing proteins with no clear function as caspase activator.
- Other.
Other indicates that a protein cannot be classified in any of the groups described above.



